KRISHNA CHOUDHARY, Ph. D.
Data Scientist | Biomedical Engineer
Unraveling the phenotypic dysregulation in complex diseases
I design and implement cutting-edge high-throughput technologies to explore the complexities of biological systems. Thus far, my research has yielded innovative wet-lab and computational technologies, and insights into systems ranging from microbes, RNA viruses, yeast to neurological and cardiovascular disease models. My pursuit of basic science includes active collaborations with diverse biomedical researchers to drive discoveries that can have an impact on human health.
I hold a Ph.D. in Biomedical Engineering from UC Davis and did my postdoctoral research at the UCSF. During my postdoc, I developed a scalable platform for imaging-based profiling of CRISPR perturbations, where I combined CRISPR technologies, multiplexed imaging, and systems biology. Now, I am continuing to apply my skills to address unmet needs in the clinic. Stay tuned for more updates!
RESEARCH
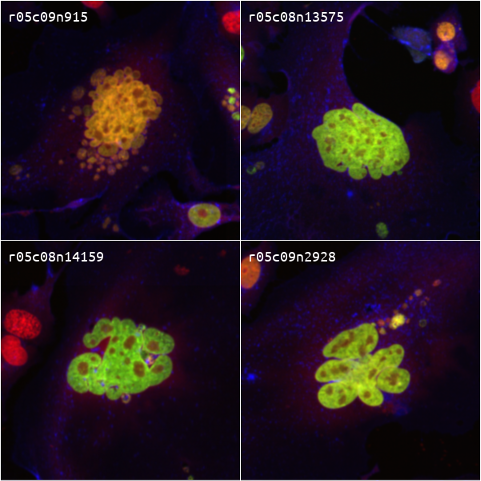 Pooled optical CRISPR screens. The large majority of human genes do not have an impact on cell proliferation. Yet, high-throughput
functional studies of genetic elements have largely focused on cell viability due to the technological challenges in large-scale measurements of
more complex phenotypes, e.g., subcellular structures and localization of proteins/RNAs. If we could characterize the phenotypic impacts of perturbing
all human genes, it could help us identify new therapies for genetic diseases. During my postdoctoral work,
I developed a cost-effective technology that scales up protein barcodes for the purpose of pooled optical CRISPR screens of
mammalian cell cultures. I applied this technology to study the
genetic networks underlying cell cycle and DNA damage.
Pooled optical CRISPR screens. The large majority of human genes do not have an impact on cell proliferation. Yet, high-throughput
functional studies of genetic elements have largely focused on cell viability due to the technological challenges in large-scale measurements of
more complex phenotypes, e.g., subcellular structures and localization of proteins/RNAs. If we could characterize the phenotypic impacts of perturbing
all human genes, it could help us identify new therapies for genetic diseases. During my postdoctoral work,
I developed a cost-effective technology that scales up protein barcodes for the purpose of pooled optical CRISPR screens of
mammalian cell cultures. I applied this technology to study the
genetic networks underlying cell cycle and DNA damage.
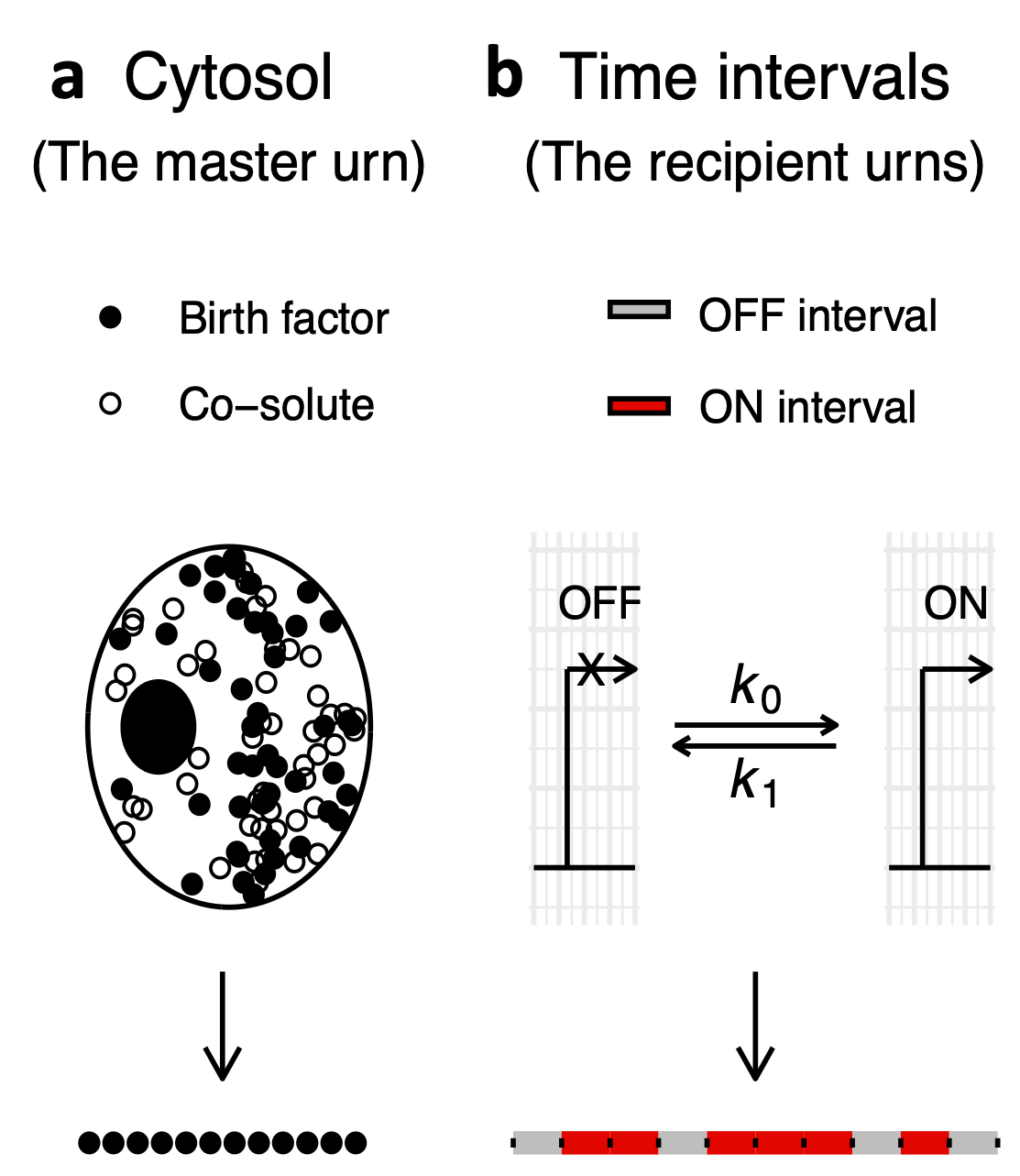 Single-cell profiling. The technological advances of the recent decades have laid bare the immense heterogeneity that exists
in molecular compositions of single cells within and between tissues. At the Gladstone Institute of Data
Science and Biotechnology, San Francisco, I powered diverse single-cell studies of cardiovascular, neurological, and infectious disease models.
On the basic science side, I developed a novel stochastic modeling framework for the probability distributions of single-cell mRNA and protein counts.
In my postdoctoral work at UCSF, I developed harmonized experimental and computational frameworks to acquire and analyze
image-based single-cell profiling data. In the future, I aim to integrate data from diverse high-throughput technologies to unravel the
mechanisms underlying human disease at the single-cell level.
Single-cell profiling. The technological advances of the recent decades have laid bare the immense heterogeneity that exists
in molecular compositions of single cells within and between tissues. At the Gladstone Institute of Data
Science and Biotechnology, San Francisco, I powered diverse single-cell studies of cardiovascular, neurological, and infectious disease models.
On the basic science side, I developed a novel stochastic modeling framework for the probability distributions of single-cell mRNA and protein counts.
In my postdoctoral work at UCSF, I developed harmonized experimental and computational frameworks to acquire and analyze
image-based single-cell profiling data. In the future, I aim to integrate data from diverse high-throughput technologies to unravel the
mechanisms underlying human disease at the single-cell level.
CONTACT
Overlapping interests? Drop a note!
Email: Krishna.Choudhary.Research@gmail.com